AbstractWith the development of more advanced methods for the diagnosis and treatment of diseases, the data required for medical care are becoming complex, and misinterpretation of information due to human error may result in serious consequences. Human error can be avoided with the support of artificial intelligence (AI). AI models trained with various medical data for diagnosis and management of liver diseases have been applied to hepatitis, fatty liver disease, liver cirrhosis, and liver cancer. Some of these models have been reported to outperform human experts in terms of performance, indicating their potential for supporting clinical practice given their high-speed output. This paper summarizes the recent advances in AI for liver disease and introduces the AI-aided diagnosis of liver tumors using B-mode ultrasonography.
IntroductionThe introduction of artificial intelligence (AI) in the medical field has been indispensable for improving the efficiency of clinical tasks and medical care with limited human resources. In clinical practice, AI is expected to process various types of data appropriately and provide necessary information to medical professionals in a form that is easily comprehensible. In other words, the purpose of medical AI is to prevent misinterpretation and support the medical decision-making process. In the field of liver disease, various AI models have been reported for several tasks, such as prediction of the most likely diagnosis and prognosis and proposals for the most suitable therapeutic approach (Fig. 1). This review introduces AI models for the diagnosis and management of liver disease. In particular, the paper reviews AI models for the ultrasonographic (US) diagnosis of liver disease [1].
AI Models Using Laboratory Data and US Images for Liver DiseaseTo date, several scoring systems for liver disease have been reported for the assessment of liver function and condition, such as the Child-Pugh score and model for end-stage liver disease (MELD) score. Since they are associated with the stage of liver disease, these scores are applicable for predicting the prognosis and outcome of treatments. Additionally, attempts have been made to evaluate the performance of AI models compared to these conventional scores [2].
AI for the Management of Hepatitis, Steatosis, and Liver CirrhosisUsing laboratory data, Ahmad et al. [3] published a model for classifying the stages of chronic hepatitis B. Another study presented an AI model using time-series laboratory data of chronic hepatitis C combined with machine learning; the results showed a good area under the receiver operating characteristic curve (AUROC) for predicting fibrosis [4-6]. Maiellaro et al. [7] developed an AI model for predicting the effect of interferon-ribavirin combination therapy using blood chemistry data of chronic hepatitis C cases. Interestingly, by applying 48,940 B-mode US images from chronic hepatitis C patients as training data in four machine learning models, Park et al. [8] reported an AI model that predicted treatment failure of direct-acting antiviral agents.
Several models have also been used to distinguish non-alcoholic steatohepatitis (NASH) from non-alcoholic fatty liver disease (NAFLD) [9]. Yip et al. [10] showed high performance of the NAFLD ridge score using blood chemistry data and a history of hypertension. In addition, some models aimed to diagnose NAFLD and NASH using large-scale medical records and lipid profiles [11-15]. Oh et al. [16] applied a random forest algorithm to analyze the gut microbiota of patients with cirrhosis associated with NAFLD, indicating that NAFLD-related cirrhosis could be identified by the microbiome. Another study reported AI for diagnosis of NAFLD using comprehensive profiling of lipids, cytokines, hormones, and metabolites [17,18].
AI can also be used to predict complications of liver cirrhosis. Marozas et al. [19] used US elastography and blood tests as training data for AI models and discriminated cases with pressure differences between the portal vein and hepatic vein. Hong et al. [20] reported a prediction model for esophageal varix emergence using the platelet count, splenomegaly, and portal vein diameter for a neural network. For patients with hepatitis C virus (HCV)–positive liver cirrhosis, AI trained with clinical data from 3,972 patients could predict the presence of esophageal varix with an accuracy of 68.9% [21]. Dong et al. [22] reported the EVendo score for the prediction of esophageal gastric varices through machine learning using blood data and ascites findings. Furthermore, as a prediction model for the prognosis of primary sclerosing cholangitis (PSC), the PSC risk estimate tool (PREsTo) was used, in which physical findings and blood chemistry data were applied as training data for gradient boosting. The PREsTo model exceeded the MELD and Mayo PSC scores for predicting liver failure [23].
Prediction of the Emergence and Management of Hepatocellular CarcinomaFor predicting the emergence of hepatocellular carcinoma (HCC) in patients with cirrhosis, the accuracy of the AI model based on the decision tree and random forest algorithms trained with time-series data from 442 patients exceeded that of conventional regression analysis [24]. A neural network-based AI model that predicted HCC emergence was reported through training time-series blood data for more than 3 years from 48,151 patients with HCV-positive cirrhosis; the results showed high performance even in patients who achieved a sustained viral response [25].
Prediction of the Outcomes of Acute Liver Failure and Liver TransplantationFor the prediction of outcomes in patients with acute liver failure, AI models based on the gradient boosting algorithm have been reported using clinical data from 527 cases, where the probability of death within 29 days after admission was presented [26]. There have also been many reports on the prediction of fibrosis of transplanted livers and outcomes after transplantation using clinical findings and laboratory data from donors and recipients [27,28]. Ayllon et al. [29] attempted to match donors and recipients using the probability of graft survival 3 months after transplantation, as indicated by a neural network. Another report presented an AI model for estimating the risk of death within 3 months among patients who were on a waiting list for brain-dead donor liver transplantation [30]. Nitski et al. [31] reported a model that predicts survival at 1 and 5 years after liver transplantation with high accuracy using AI based on transfer learning. Furthermore, AI models for predicting tumor recurrence after liver transplantation in HCC cases have also been reported [32,33].
AI for US Diagnosis of Liver DiseaseThe interpretation and diagnosis of a large number of medical images are a challenge for medical professionals; this task can be affected by human error, especially under strict time constraints. Many AI models using medical images, including US, have been reported for the diagnosis of liver disease, and some studies aimed to predict the outcome of diseases from imaging data.
AI Models for Liver TumorsUS is a noninvasive imaging modality commonly used for diagnosis of liver disease in many medical facilities. However, extensive skills and experience are required for an accurate diagnosis under time constraints. Therefore, the development of AI for US diagnosis is an attractive possibility. There have been many reports on the diagnosis of liver tumors by AI using B-mode US images; the early reports were mostly retrospective studies, with small sample sizes and unverified with an external cohort [1,34].
Hwang et al. [35] reported a model with 96% accuracy for the classification of cysts, hemangiomas, and malignant tumors. Hassan et al. [36] showed that the accuracy of four-class discrimination of AI for normal liver, cyst, hemangioma, and HCC was 97.2%. Another study reported an AI model with 95% accuracy when classifying four types of liver masses: cysts, hemangiomas, HCC, and metastatic cancers [37]. Schmauch et al. [38] reported a deep neural network-based AI by adjusting the B-mode image intensity using the US intensity of the abdominal wall to standardize the difference in image conditions. They reported an AUROC of 0.9 or higher in tumor detection and 5-class classification (HCC, metastatic tumor, hemangioma, cyst, focal nodular hyperplasia) [38].
Meanwhile, Zhang et al. [39] developed a model trained with a small number of contrast-enhanced ultrasound (CEUS) images using transfer learning of B-mode US. In addition, an AI model with an accuracy of 96.3% and AUROC of 0.994 was reported for benign and malignant tumor discrimination through combined training with B-mode US images and other information from medical records and blood data [40].
As a more practical model for detecting liver tumors, Yang et al. [41] created a convolutional neural network (CNN) model using 24,343 B-mode US images of liver tumors from 2,143 cases and validated the AI model’s performance with an independent test cohort. This model showed considerable accuracy in the diagnosis of malignant tumors, and comparable performance to that of contrast-enhanced computed tomography (CT) and magnetic resonance imaging (MRI) diagnoses by a radiologist. In that study, three models were created: an AI model trained with tumor US images, a model trained with tumor and background liver US images, and a model trained with patient backgrounds in addition to US images. The performance was particularly high when both US images and patient backgrounds were used for training. Interestingly, the ability to discriminate between benign and malignant tumors was analyzed by stratification according to tumor size, and the size of the tumor did not significantly affect the performance. In contrast, the model trained with US images and clinical data showed the best performance for discriminating benign and malignant tumors. It is notable that this AI model can output a color map in which the color tone of image pixels changes according to the degree that it contributes to the diagnosis [41].
Dadoun et al. [42] reported an AI model trained using 2,551 B-mode US images of liver tumors from 1,026 cases and verified them in an external test cohort with 155 images from 48 cases. Using a detection transformer (DETR), this AI model showed 97% sensitivity and 90% specificity for the identification of US images showing liver tumors, with a positive predictive value of 77% and sensitivity of 84% for analyzing the localization of tumors, with a mean intersection over union of 0.69. This performance was almost equivalent to that of the two experts, and the sensitivity and specificity for distinguishing malignant from benign lesions were 82% and 81%, respectively, which were higher than those of the experts. In that report, six types of tumors (HCC, metastatic liver cancer, hemangiomas, focal nodular hyperplasia, hepatic adenomas, and cysts) were classified with an overall accuracy of 76%. The performance of DETR met or exceeded those of two experts and CNN-based models for these tasks [42].
In addition to B-mode examinations, CEUS is applied for the diagnosis of liver tumors, and a multi-kernel AI learning model for discriminating malignant tumors from benign lesions has been reported [43]. Furthermore, another report discriminated atypical HCC from focal nodular hyperplasia through an AI system pretrained with CEUS images; this type of discrimination is difficult for humans [44].
AI Models for Diffuse Liver LesionsMany reported AI models have focused on classifying the stage of fibrosis and steatosis for the diagnosis of diffuse liver lesions [1]. A model where B-mode US images were applied for training a neural network showed an accuracy of 97.3%, sensitivity of 96%, and specificity of 100% for discrimination of normal liver, fatty liver, and cirrhosis [45]. Another report showed an AI model for estimating the presence of liver cirrhosis, portal hypertension, and esophageal varices through training a support vector machine (SVM) with images of shear wave elastography (SWE) [46,47]. A study also reported the use of AI for discriminating the stage of liver fibrosis by applying Doppler US parameters as training data [48]. Wang et al. [49] established a deep learning model using 1,990 images of SWE from 398 hepatitis B virus (HBV)–positive patients as training data; its performance for discriminating the fibrosis stage was superior to those of 2D-SWE, the aspartate aminotransferase to platelet ratio index (APRI), and the fibrosis-4 (FIB-4) index. AI models for discriminating the fibrosis stage using multiple B-mode and CEUS parameters have been reported as well [50].
Development and Perspective of B-mode US Diagnosis Support Model for Focal Liver LesionsTo accumulate training data for B-mode US diagnosis by AI, the authors have constructed a US image database. Based on these datasets, the development of AI models that support the diagnosis of liver tumors in B-mode US has been initiated. In addition, with a view to social implementation of the developed AI, evaluation tests for its performance are being conducted.
An AI system has been developed to assist in the diagnosis of focal liver lesions using B-mode US (Fig. 2) [51]. A total of 83,569 images of four types of liver masses (HCC, metastatic cancer, hemangiomas, and cysts) were used for training the 19-layer CNN. In the evaluation of the 10-fold cross-validation, the differential accuracy for the four classes of diseases was 90.8%, and the sensitivities for diagnosing HCC, metastatic cancer, hemangiomas, and cysts were 73.8%, 61.7%, 89.8%, and 99.1%, respectively. The accuracy, sensitivity, and specificity of malignancy discrimination were 94.2%, 85.6%, and 96.2%, respectively. Prior to this, AI models using 7,754, 24,575, 57,145, and 70,950 images for training were created, and the accuracy of four-class and malignant tumor discrimination improved according to the amount of training data (Fig. 3) [51].
The diagnostic capabilities of AI and human experts were also compared in an independent test cohort of tumor video images using a 70,950-image learning model (Fig. 4). For AI, the accuracy of the four-disease classification was 89.1% and for the diagnosis of malignant tumors was 90.9%. Meanwhile, the median accuracy of the four-disease classification by human experts was 67.3% (range, 63.6% to 69.1%), and for the diagnosis of malignant tumors by human experts, it was 80.0% (range, 74.5% to 83.6%). In addition, the probability of AI making the correct diagnosis increases with a larger amount of training data, indicating that a highly reliable estimation can be acquired by increasing the amount of training data [51].
AI for Other Imaging ModalitiesAI Models for CT and MRIGenerally, a further examination is required for a definitive diagnosis of the lesions detected on US, and CT and MRI are recommended for this purpose [52]. Yasaka et al. [53] reported that an CNN-based AI model pretrained with CT images of 460 liver tumors for the diagnosis of malignant tumors had an accuracy of 84% and an AUROC of 0.927. Nayak et al. [54] performed three-dimensional image segmentation of the liver region and HCC from CT images using deep learning and reported that the prediction accuracy for each cross-section was 86.9% and the prediction accuracy for each case was 80%. Hamm et al. [55] used 434 MRI images as training data for a CNN to create an AI-aided detection and diagnosis model for HCC, which outperformed human radiologists. Feng et al. [56] used gadolinium ethoxybenzyl diethlenetriamine pentaacetic acid (Gd-EOB-DTPA)-MRI images to preoperatively estimate microvascular invasion in HCC cases based on the pathological findings of resected specimens, which reportedly surpassed the diagnosis of the radiologist. For diffuse hepatic lesions, Choi et al. [57] reported a CNN model for diagnosing the fibrosis stage using 7,461 CT images as training data, showing superior accuracy compared to that of radiologists, APRI, and FIB-4 index. In addition, several SVM-based AI models predicting liver fibrosis and liver stiffness based on MRI have been reported [58,59].
AI for Pathological Diagnosis of Liver LesionsAI is also expected to support the pathological diagnosis of whole-slide images (WSIs), which is required for a definitive diagnosis of lesions. A CNN model for classifying nuclear atypia of HCC on biopsy was reported [60]. Liao et al. [61] used WSIs of hematoxylin and eosin (H&E) staining and tissue microarray to perform an automatic diagnosis of HCC and predict gene mutations. They showed that CTNNB1 (β-catenin) gene mutations could be predicted from pathological specimens of H&E staining. Chen et al. [62] reported a neural network for benign and malignant tumor discrimination with an accuracy of 96.0% and classification of well, moderately, and poorly differentiated HCC with an accuracy of 86.9% through training on WSIs of HCC and non-cancerous liver. They showed that the performance of AI was equivalent to that of a pathologist with 5 years of experience. They also reported that gene mutations from H&E specimens could be predicted using AI.
AI for Predicting the Prognosis in HCCBecause the selection of the best treatment based on the estimated prognosis is critical for the management of HCC, the outcome of the treatment is sometimes rather unpredictable in each HCC case. Therefore, several AIs trained with image and omics data have been reported for predicting the prognosis after the treatment.
Regarding AI based on CEUS images, deep learning radiomics models have been reported, which may optimize the treatments for very early/early-stage HCC. The models identified that 17.3% of HCC patients who underwent radiofrequency ablation and 27.3% of patients who underwent surgical resection should swap their treatment, so that their average probability of 2-year progression-free survival would increase by 12% and 15%, respectively [63]. Another report presented an AI model that predicted the effect of transcatheter chemoembolization (TACE) by analyzing video images of contrast medium inflow into tumors on CEUS [64].
Using EOB-MRI images, Kim et al. [65] reported an AI model based on a random forest algorithm to predict postoperative recurrence-free survival in solitary HCC cases with diameters of 2 to 5 cm. For predicting the early recurrence of HCC, it has been reported that an AI model pretrained with tumor images of EOB-MRI, including up to 3 mm outside the tumor margin, showed the same performance as a model trained with pathological images of HCC. CNN-based AI models trained with CT images of HCC before treatment and a random forest-based AI trained with MRI images reportedly predicted tumor response to TACE [66,67].
However, Saillard et al. [68] reported a CNN model that predicts postoperative survival time by learning the WSI of resected specimens. Using WSI and omics data from HCC cases, Shi et al. [69] reported a model that presented a "tumor risk score," which was associated with survival. Survival prediction based on this score is superior to known conventional scores and associated with a specific tumor immune environment and genetic mutations. Chaudhary et al. [70] developed a deep neural network model and found two groups with different prognoses using RNA sequencing, microRNA sequencing, and methylation data from 360 patients with HCC.
ConclusionA variety of AI models for the diagnosis of liver disease have been reported; some of them reportedly outperform human experts. However, the value of the output from AI could be more informative for beginners and non-experts because AI generally shows much higher performance than beginners and non-experts, compared to experts, especially in the field of imaging diagnosis [42,51]. Another study indicated that AI for diagnosing focal liver lesions in B-mode US shows the potential to assist less-experienced radiologists in improving their performance and lowering their dependence on sectional imaging in liver cancer diagnosis [41], although medical staff bear the final responsibility for medical decisions.
However, the use of AI may still be limited in the clinical setting; the performance of AI is completely dependent on the quantity and quality of training data. For example, regarding AI for image diagnosis, an imbalance in the distribution of diseases among the training images would lead to a biased output—that is, a preferential diagnosis of the disease most contained in the training data set [2,37]. Therefore, the performance of AI may fluctuate among the test cohorts, and physicians must be careful about the value of output by the AI until the results of AI studies are proven through clinical trials. In this context, it is becoming important to understand the advantages and disadvantages of medical AI in clinical settings.
NotesAuthor Contributions Conceptualization: Nishida N, Kudo M. Data acquisition: Nishida N. Data analysis or interpretation: Nishida N. Drafting of the manuscript: Nishida N. Critical revision of the manuscript: Nishida N, Kudo M. Approval of the final version of the manuscript: all authors. AcknowledgementsAI development of the Japan Society of Ultrasonics in Medicine is carried out with the support of AMED (JP18lk1010030, JP20lk1010035), and the tumor discriminator is being developed under the initiative of Dr. Makoto Yamakawa and Professor Takeshi Shiina, Kyoto University Graduate School of Medicine, Department of Human Health Sciences. The tumor detector is being developed under the initiative of Professor Yoshito Mekada, Graduate School of Engineering, Chukyo University. We would also like to take this opportunity to thank the members of the Japan Society of Ultrasonics in Medicine for their cooperation in carrying out the research.
References1. Nishida N, Yamakawa M, Shiina T, Kudo M. Current status and perspectives for computer-aided ultrasonic diagnosis of liver lesions using deep learning technology. Hepatol Int 2019;13:416–421.
2. Ahn JC, Connell A, Simonetto DA, Hughes C, Shah VH. Application of artificial intelligence for the diagnosis and treatment of liver diseases. Hepatology 2021;73:2546–2563.
3. Ahmad G, Khan MA, Abbas S, Athar A, Khan BS, Aslam MS. Automated diagnosis of hepatitis B using Multilayer Mamdani Fuzzy Inference System. J Healthc Eng 2019;2019:6361318.
4. Konerman MA, Zhang Y, Zhu J, Higgins PD, Lok AS, Waljee AK. Improvement of predictive models of risk of disease progression in chronic hepatitis C by incorporating longitudinal data. Hepatology 2015;61:1832–1841.
5. Konerman MA, Lu D, Zhang Y, Thomson M, Zhu J, Verma A, et al. Assessing risk of fibrosis progression and liver-related clinical outcomes among patients with both early stage and advanced chronic hepatitis C. PLoS One 2017;12:e0187344.
6. Konerman MA, Beste LA, Van T, Liu B, Zhang X, Zhu J, et al. Machine learning models to predict disease progression among veterans with hepatitis C virus. PLoS One 2019;14:e0208141.
7. Maiellaro PA, Cozzolongo R, Marino P. Artificial neural networks for the prediction of response to interferon plus ribavirin treatment in patients with chronic hepatitis C. Curr Pharm Des 2004;10:2101–2109.
8. Park H, Lo-Ciganic WH, Huang J, Wu Y, Henry L, Peter J, et al. Machine learning algorithms for predicting direct-acting antiviral treatment failure in chronic hepatitis C: an HCV-TARGET analysis. Hepatology 2022;76:483–491.
9. Decharatanachart P, Chaiteerakij R, Tiyarattanachai T, Treeprasertsuk S. Application of artificial intelligence in non-alcoholic fatty liver disease and liver fibrosis: a systematic review and meta-analysis. Therap Adv Gastroenterol 2021;14:17562848211062807.
10. Yip TC, Ma AJ, Wong VW, Tse YK, Chan HL, Yuen PC, et al. Laboratory parameter-based machine learning model for excluding non-alcoholic fatty liver disease (NAFLD) in the general population. Aliment Pharmacol Ther 2017;46:447–456.
11. Islam MM, Wu CC, Poly TN, Yang HC, Li YJ. Applications of machine learning in fatty live disease prediction. Stud Health Technol Inform 2018;247:166–170.
12. Ma H, Xu CF, Shen Z, Yu CH, Li YM. Application of machine learning techniques for clinical predictive modeling: a cross-sectional study on nonalcoholic fatty liver disease in China. Biomed Res Int 2018;2018:4304376.
13. Wu CC, Yeh WC, Hsu WD, Islam MM, Nguyen PA, Poly TN, et al. Prediction of fatty liver disease using machine learning algorithms. Comput Methods Programs Biomed 2019;170:23–29.
14. Perveen S, Shahbaz M, Keshavjee K, Guergachi A. A systematic machine learning based approach for the diagnosis of non-alcoholic fatty liver disease risk and progression. Sci Rep 2018;8:2112.
15. Okanoue T, Shima T, Mitsumoto Y, Umemura A, Yamaguchi K, Itoh Y, et al. Artificial intelligence/neural network system for the screening of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis. Hepatol Res 2021;51:554–569.
16. Oh TG, Kim SM, Caussy C, Fu T, Guo J, Bassirian S, et al. A universal gut-microbiome-derived signature predicts cirrhosis. Cell Metab 2020;32:901.
17. Sowa JP, Atmaca O, Kahraman A, Schlattjan M, Lindner M, Sydor S, et al. Non-invasive separation of alcoholic and non-alcoholic liver disease with predictive modeling. PLoS One 2014;9:e101444.
18. Perakakis N, Polyzos SA, Yazdani A, Sala-Vila A, Kountouras J, Anastasilakis AD, et al. Non-invasive diagnosis of non-alcoholic steatohepatitis and fibrosis with the use of omics and supervised learning: a proof of concept study. Metabolism 2019;101:154005.
19. Marozas M, Zykus R, Sakalauskas A, Kupcinskas L, Lukosevicius A. Noninvasive evaluation of portal hypertension using a supervised learning technique. J Healthc Eng 2017;2017:6183714.
20. Hong WD, Zhu QH, Huang ZM, Chen XR, Jiang ZC, Xu SH, et al. Predictors of esophageal varices in patients with HBV-related cirrhosis: a retrospective study. BMC Gastroenterol 2009;9:11.
21. Abd El-Salam SM, Ezz MM, Hashem S, Elakel W, Salama R, ElMakhzangy H, et al. Performance of machine learning approaches on prediction of esophageal varices for Egyptian chronic hepatitis C patients. Informa Med Unlocked 2019;17:100267.
22. Dong TS, Kalani A, Aby ES, Le L, Luu K, Hauer M, et al. Machine learning-based development and validation of a scoring system for screening high-risk esophageal varices. Clin Gastroenterol Hepatol 2019;17:1894–1901.
23. Eaton JE, Vesterhus M, McCauley BM, Atkinson EJ, Schlicht EM, Juran BD, et al. Primary Sclerosing Cholangitis Risk Estimate Tool (PREsTo) predicts outcomes of the disease: a derivation and validation study using machine learning. Hepatology 2020;71:214–224.
24. Singal AG, Mukherjee A, Elmunzer BJ, Higgins PD, Lok AS, Zhu J, et al. Machine learning algorithms outperform conventional regression models in predicting development of hepatocellular carcinoma. Am J Gastroenterol 2013;108:1723–1730.
25. Ioannou GN, Tang W, Beste LA, Tincopa MA, Su GL, Van T, et al. Assessment of a deep learning model to predict hepatocellular carcinoma in patients with hepatitis C Cirrhosis. JAMA Netw Open 2020;3:e2015626.
26. Garcia MS, Agarwal B, Mookerjee RP, Jalan R, Doyle G, Ranco G, et al. An accurate data preparation approach for the prediction of mortality in ACLF patients using the CANONIC dataset. Annu Int Conf IEEE Eng Med Biol Soc 2019;2019:1371–1377.
27. Piscaglia F, Cucchetti A, Benlloch S, Vivarelli M, Berenguer J, Bolondi L, et al. Prediction of significant fibrosis in hepatitis C virus infected liver transplant recipients by artificial neural network analysis of clinical factors. Eur J Gastroenterol Hepatol 2006;18:1255–1261.
28. Briceno J, Cruz-Ramirez M, Prieto M, Navasa M, Ortiz de Urbina J, Orti R, et al. Use of artificial intelligence as an innovative donor-recipient matching model for liver transplantation: results from a multicenter Spanish study. J Hepatol 2014;61:1020–1028.
29. Ayllon MD, Ciria R, Cruz-Ramirez M, Perez-Ortiz M, Gomez I, Valente R, et al. Validation of artificial neural networks as a methodology for donor-recipient matching for liver transplantation. Liver Transpl 2018;24:192–203.
30. Bertsimas D, Kung J, Trichakis N, Wang Y, Hirose R, Vagefi PA. Development and validation of an optimized prediction of mortality for candidates awaiting liver transplantation. Am J Transplant 2019;19:1109–1118.
31. Nitski O, Azhie A, Qazi-Arisar FA, Wang X, Ma S, Lilly L, et al. Longterm mortality risk stratification of liver transplant recipients: realtime application of deep learning algorithms on longitudinal data. Lancet Digit Health 2021;3:e295–e305.
32. Nam JY, Lee JH, Bae J, Chang Y, Cho Y, Sinn DH, et al. Novel model to predict HCC recurrence after liver transplantation obtained using deep learning: a multicenter study. Cancers (Basel) 2020;12:2791.
33. Ivanics T, Nelson W, Patel MS, Claasen M, Lau L, Gorgen A, et al. The Toronto postliver transplantation hepatocellular carcinoma recurrence calculator: a machine learning approach. Liver Transpl 2022;28:593–602.
34. Nishida N, Kudo M. Artificial intelligence in medical imaging and its application in sonography for the management of liver tumor. Front Oncol 2020;10:594580.
35. Hwang YN, Lee JH, Kim GY, Jiang YY, Kim SM. Classification of focal liver lesions on ultrasound images by extracting hybrid textural features and using an artificial neural network. Biomed Mater Eng 2015;26 Suppl 1:S1599–S1611.
36. Hassan TM, Elmogy M, Salam ES. Diagnosis of focal liver diseases based on deep learning technique for ultrasound images. Arab J Sci Eng 2017;42:3127–3140.
37. Virmani J, Kumar V, Kalra N, Khandelwal N. Neural network ensemble based CAD system for focal liver lesions from B-mode ultrasound. J Digit Imaging 2014;27:520–537.
38. Schmauch B, Herent P, Jehanno P, Dehaene O, Saillard C, Aube C, et al. Diagnosis of focal liver lesions from ultrasound using deep learning. Diagn Interv Imaging 2019;100:227–233.
39. Zhang H, Guo L, Wang D, Wang J, Bao L, Ying S, et al. Multi-source transfer learning via multi-kernel support vector machine plus for B-mode ultrasound-based computer-aided diagnosis of liver cancers. IEEE J Biomed Health Inform 2021;25:3874–3885.
40. Sato M, Kobayashi T, Soroida Y, Tanaka T, Nakatsuka T, Nakagawa H, et al. Development of novel deep multimodal representation learning-based model for the differentiation of liver tumors on B-mode ultrasound images. J Gastroenterol Hepatol 2022;37:678–684.
41. Yang Q, Wei J, Hao X, Kong D, Yu X, Jiang T, et al. Improving B-mode ultrasound diagnostic performance for focal liver lesions using deep learning: a multicentre study. EBioMedicine 2020;56:102777.
42. Dadoun H, Rousseau AL, de Kerviler E, Correas JM, Tissier AM, Joujou F, et al. Deep learning for the detection, localization, and characterization of focal liver lesions on abdominal US images. Radiol Artif Intell 2022;4:e210110.
43. Guo LH, Wang D, Qian YY, Zheng X, Zhao CK, Li XL, et al. A twostage multi-view learning framework based computer-aided diagnosis of liver tumors with contrast enhanced ultrasound images. Clin Hemorheol Microcirc 2018;69:343–354.
44. Li W, Lv XZ, Zheng X, Ruan SM, Hu HT, Chen LD, et al. Machine learning-based ultrasomics improves the diagnostic performance in differentiating focal nodular hyperplasia and atypical hepatocellular carcinoma. Front Oncol 2021;11:544979.
45. Acharya UR, Raghavendra U, Fujita H, Hagiwara Y, Koh JE, Jen Hong T, et al. Automated characterization of fatty liver disease and cirrhosis using curvelet transform and entropy features extracted from ultrasound images. Comput Biol Med 2016;79:250–258.
46. Procopet B, Cristea VM, Robic MA, Grigorescu M, Agachi PS, Metivier S, et al. Serum tests, liver stiffness and artificial neural networks for diagnosing cirrhosis and portal hypertension. Dig Liver Dis 2015;47:411–416.
47. Gatos I, Tsantis S, Spiliopoulos S, Karnabatidis D, Theotokas I, Zoumpoulis P, et al. A machine-learning algorithm toward color analysis for chronic liver disease classification, employing ultrasound shear wave elastography. Ultrasound Med Biol 2017;43:1797–1810.
48. Zhang L, Li QY, Duan YY, Yan GZ, Yang YL, Yang RJ. Artificial neural network aided non-invasive grading evaluation of hepatic fibrosis by duplex ultrasonography. BMC Med Inform Decis Mak 2012;12:55.
49. Wang K, Lu X, Zhou H, Gao Y, Zheng J, Tong M, et al. Deep learning radiomics of shear wave elastography significantly improved diagnostic performance for assessing liver fibrosis in chronic hepatitis B: a prospective multicentre study. Gut 2019;68:729–741.
50. Li W, Huang Y, Zhuang BW, Liu GJ, Hu HT, Li X, et al. Multiparametric ultrasomics of significant liver fibrosis: a machine learning-based analysis. Eur Radiol 2019;29:1496–1506.
51. Nishida N, Yamakawa M, Shiina T, Mekada Y, Nishida M, Sakamoto N, et al. Artificial intelligence (AI) models for the ultrasonographic diagnosis of liver tumors and comparison of diagnostic accuracies between AI and human experts. J Gastroenterol 2022;57:309–321.
52. Kokudo N, Takemura N, Hasegawa K, Takayama T, Kubo S, Shimada M, et al. Clinical practice guidelines for hepatocellular carcinoma: the Japan Society of Hepatology 2017 (4th JSH-HCC guidelines) 2019 update. Hepatol Res 2019;49:1109–1113.
53. Yasaka K, Akai H, Abe O, Kiryu S. Deep learning with vonvolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 2018;286:887–896.
54. Nayak A, Baidya Kayal E, Arya M, Culli J, Krishan S, Agarwal S, et al. Computer-aided diagnosis of cirrhosis and hepatocellular carcinoma using multi-phase abdomen CT. Int J Comput Assist Radiol Surg 2019;14:1341–1352.
55. Hamm CA, Wang CJ, Savic LJ, Ferrante M, Schobert I, Schlachter T, et al. Deep learning for liver tumor diagnosis part I: development of a convolutional neural network classifier for multi-phasic MRI. Eur Radiol 2019;29:3338–3347.
56. Feng ST, Jia Y, Liao B, Huang B, Zhou Q, Li X, et al. Preoperative prediction of microvascular invasion in hepatocellular cancer: a radiomics model using Gd-EOB-DTPA-enhanced MRI. Eur Radiol 2019;29:4648–4659.
57. Choi KJ, Jang JK, Lee SS, Sung YS, Shim WH, Kim HS, et al. Development and validation of a deep learning system for staging liver fibrosis by using contrast agent-enhanced CT images in the liver. Radiology 2018;289:688–697.
58. He L, Li H, Dudley JA, Maloney TC, Brady SL, Somasundaram E, et al. Machine learning prediction of liver stiffness using clinical and T2-weighted MRI radiomic data. AJR Am J Roentgenol 2019;213:592–601.
59. Ahmed Y, Hussein RS, Basha TA, Khalifa AM, Ibrahim AS, Abdelmoaty AS, et al. Detecting liver fibrosis using a machine learning-based approach to the quantification of the heart-induced deformation in tagged MR images. NMR Biomed 2020;33:e4215.
60. Li S, Jiang H, Pang W. Joint multiple fully connected convolutional neural network with extreme learning machine for hepatocellular carcinoma nuclei grading. Comput Biol Med 2017;84:156–167.
61. Liao H, Long Y, Han R, Wang W, Xu L, Liao M, et al. Deep learningbased classification and mutation prediction from histopathological images of hepatocellular carcinoma. Clin Transl Med 2020;10:e102.
62. Chen M, Zhang B, Topatana W, Cao J, Zhu H, Juengpanich S, et al. Classification and mutation prediction based on histopathology H&E images in liver cancer using deep learning. NPJ Precis Oncol 2020;4:14.
63. Liu F, Liu D, Wang K, Xie X, Su L, Kuang M, et al. Deep learning radiomics based on contrast-enhanced ultrasound might optimize curative treatments for very-early or early-stage hepatocellular carcinoma patients. Liver Cancer 2020;9:397–413.
64. Liu D, Liu F, Xie X, Su L, Liu M, Xie X, et al. Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrastenhanced ultrasound. Eur Radiol 2020;30:2365–2376.
65. Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY. Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res 2019;25:3847–3855.
66. Abajian A, Murali N, Savic LJ, Laage-Gaupp FM, Nezami N, Duncan JS, et al. Predicting treatment response to intra-arterial therapies for hepatocellular carcinoma with the use of supervised machine learning: an artificial intelligence concept. J Vasc Interv Radiol 2018;29:850–857.
67. Morshid A, Elsayes KM, Khalaf AM, Elmohr MM, Yu J, Kaseb AO, et al. A machine learning model to predict hepatocellular carcinoma response to transcatheter arterial chemoembolization. Radiol Artif Intell 2019;1:e180021.
68. Saillard C, Schmauch B, Laifa O, Moarii M, Toldo S, Zaslavskiy M, et al. Predicting survival after hepatocellular carcinoma resection using deep learning on histological slides. Hepatology 2020;72:2000–2013.
Role of medical artificial intelligence (AI) in the management of liver disease.Abundant medical information is accumulated in the healthcare field, such as physical findings, blood chemical data, medical images, and clinical reports. However, it is sometimes difficult to obtain critical information from a large amount of data instantly. Medical AI is expected to process information from medical records and to provide important information to medical staff in a form that is easily comprehensible. In the area of liver disease, there are many reports of AI for the diagnosis and management of hepatitis, fatty liver, cirrhosis, hepatocellular carcinoma, liver failure, and liver transplantation. CT, computed tomography; MRI, magnetic resonance imaging.
 Fig. 1.Artificial intelligence (AI) for four-class discrimination of liver masses.A still image of B-mode ultrasonography shows an iso-high-echoic mass on the surface of the liver parenchyma, which is indicated by the white arrows (A). Display of AI shows the four-class discrimination of liver masses, including cyst, hemangioma, hepatocellular carcinoma (HCC) and metastatic tumor (metastatic) with their diagnostic probabilities (B). After setting the boundary box on this liver mass (indicated as a red box), the AI automatically presents the estimated probability for the diagnosis of each type of liver mass, where the probability for hemangioma is the highest (79.0%, shown in the bottom right corner).
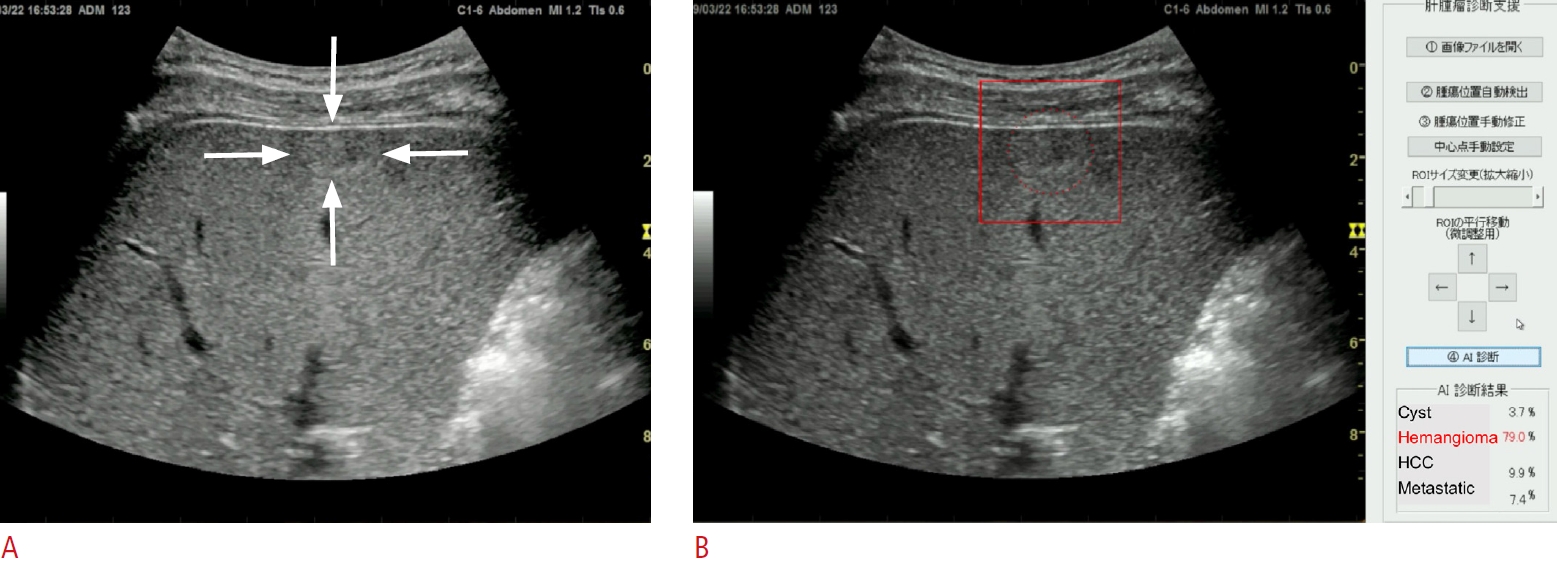 Fig. 2.Improvement of the diagnostic accuracy of artificial intelligence (AI) for the classification of liver masses according to the increase of training data.The diagnostic accuracy of AI that differentiates the four types of liver masses (hepatocellular carcinoma, metastatic tumor, hemangioma, and cyst) is shown (A). The accuracies are calculated using 10-fold cross-validation. The accuracy for four-disease classification and those for each type of liver mass improve with an increasing number of images applied for training the AI. Similarly, the accuracy, sensitivity, and specificity for the discrimination of malignant tumors also improve with an increasing number of training images (B).
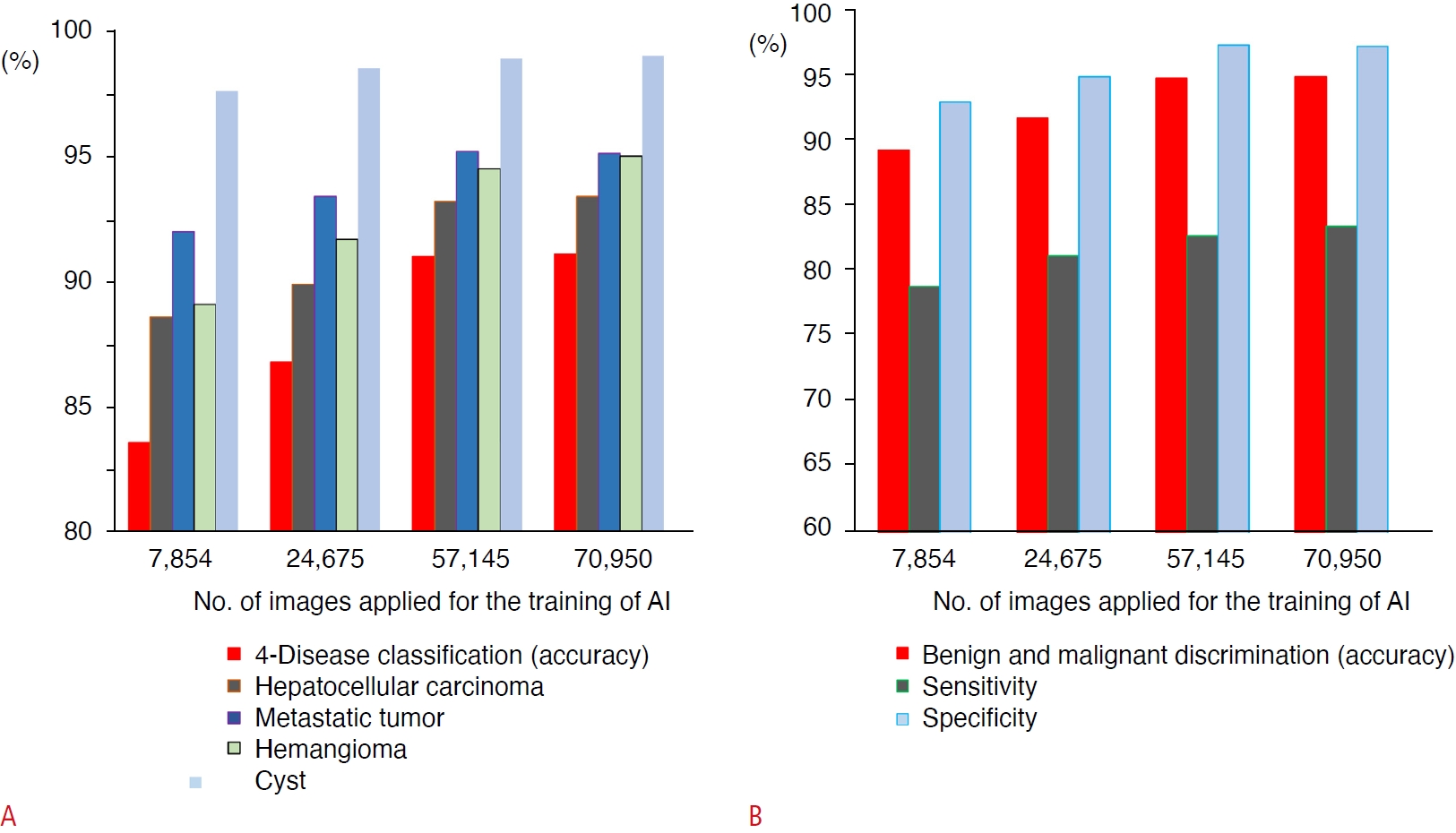 Fig. 3.Comparison of diagnostic accuracy between artificial intelligence (AI) models and physicians in an independent test cohort of focal liver lesions.The accuracy of the four-disease classification by AI was 89.1% in a learning model with 70,950 images, 81.8% in a learning model with 57,145 images, and 80.0% in a learning model with 24,575 images, while the median percentages of diagnostic accuracy by physicians were 67.3% (range, 63.6% to 69.1%) and 47.3% (range, 45.5% to 47.3%) for experts and non-experts, respectively (A). Similarly, the accuracy for discrimination of malignant tumors by AI was 90.9% in the learning model with 70,950 images, and 87.3% in the learning models with 57,145 and 24,575 images, respectively. For the human physicians, the median the diagnostic accuracy among the test nodules was 80.0% (range, 74.5% to 83.6%) by experts and 60.0% (range, 52.7% to 65.5%) by non-experts, respectively (B). Modified from Nishida N et al. J Gastroenterol 2022;57:309-321 [51], according to Creative Commons licence.
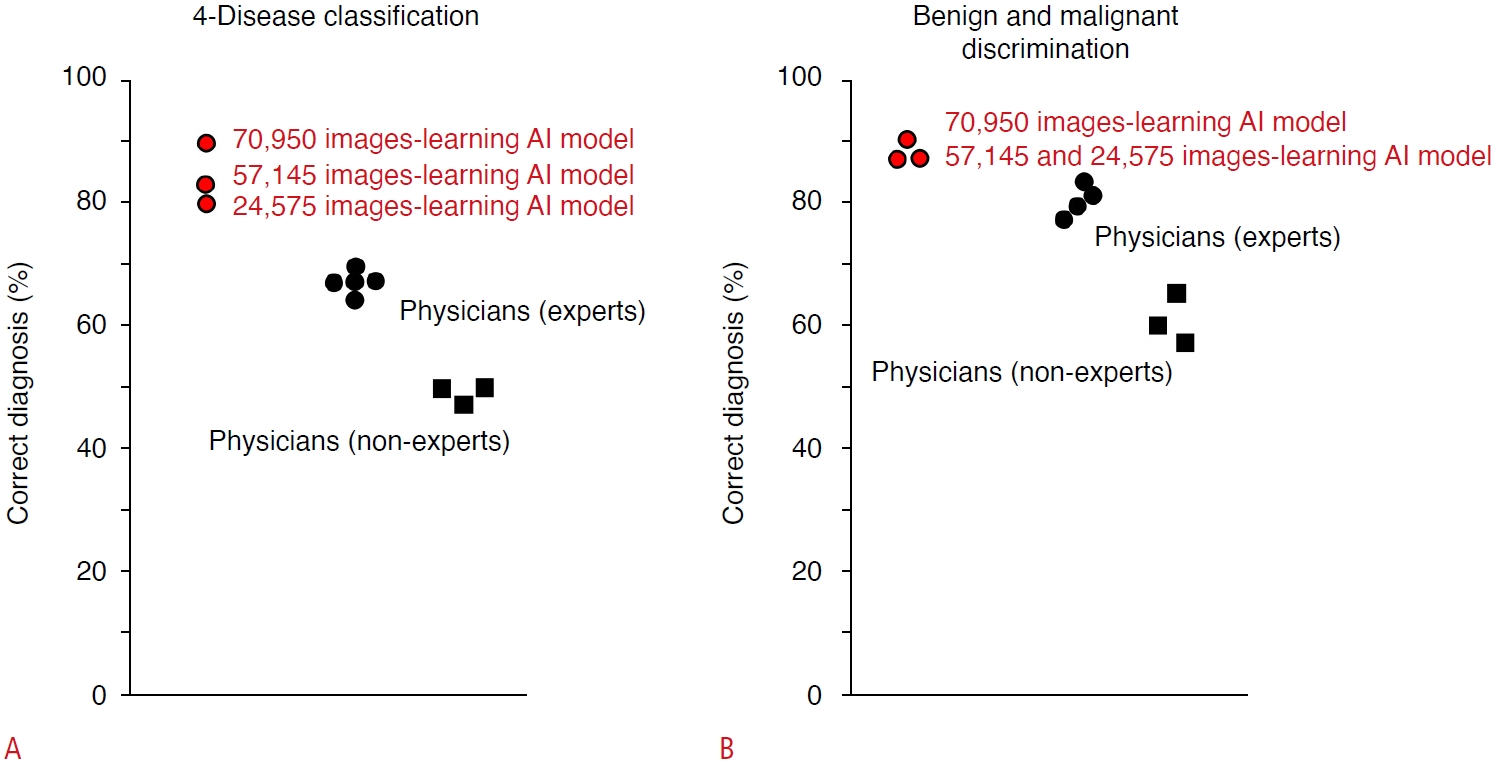 Fig. 4. |